23. Neural Network Regression with JAX#
GPU
This lecture was built using a machine with access to a GPU.
Google Colab has a free tier with GPUs that you can access as follows:
Click on the “play” icon top right
Select Colab
Set the runtime environment to include a GPU
23.1. Outline#
In a previous lecture, we showed how to implement regression using a neural network via the deep learning library Keras.
In this lecture, we solve the same problem directly, using JAX operations rather than relying on the Keras frontend.
The objectives are
Understand the nuts and bolts of the exercise better
Explore more features of JAX
Observe how using JAX directly allows us to greatly improve performance.
The lecture proceeds in three stages:
We solve the problem using Keras, to give ourselves a benchmark.
We solve the same problem in pure JAX, using pytree operations and gradient descent.
We solve the same problem using a combination of JAX and Optax, an optimization library built for JAX.
We begin with imports and installs.
import numpy as np
import jax
import jax.numpy as jnp
import matplotlib.pyplot as plt
import os
from time import time
from typing import NamedTuple
from functools import partial
!pip install keras optax
Show code cell output
Requirement already satisfied: keras in /usr/local/lib/python3.12/dist-packages (3.10.0)
Requirement already satisfied: optax in /usr/local/lib/python3.12/dist-packages (0.2.6)
Requirement already satisfied: absl-py in /usr/local/lib/python3.12/dist-packages (from keras) (1.4.0)
Requirement already satisfied: numpy in /usr/local/lib/python3.12/dist-packages (from keras) (2.0.2)
Requirement already satisfied: rich in /usr/local/lib/python3.12/dist-packages (from keras) (13.9.4)
Requirement already satisfied: namex in /usr/local/lib/python3.12/dist-packages (from keras) (0.1.0)
Requirement already satisfied: h5py in /usr/local/lib/python3.12/dist-packages (from keras) (3.15.1)
Requirement already satisfied: optree in /usr/local/lib/python3.12/dist-packages (from keras) (0.17.0)
Requirement already satisfied: ml-dtypes in /usr/local/lib/python3.12/dist-packages (from keras) (0.5.3)
Requirement already satisfied: packaging in /usr/local/lib/python3.12/dist-packages (from keras) (25.0)
Requirement already satisfied: chex>=0.1.87 in /usr/local/lib/python3.12/dist-packages (from optax) (0.1.90)
Requirement already satisfied: jax>=0.5.3 in /usr/local/lib/python3.12/dist-packages (from optax) (0.7.2)
Requirement already satisfied: jaxlib>=0.5.3 in /usr/local/lib/python3.12/dist-packages (from optax) (0.7.2)
Requirement already satisfied: typing_extensions>=4.2.0 in /usr/local/lib/python3.12/dist-packages (from chex>=0.1.87->optax) (4.15.0)
Requirement already satisfied: setuptools in /usr/local/lib/python3.12/dist-packages (from chex>=0.1.87->optax) (75.2.0)
Requirement already satisfied: toolz>=0.9.0 in /usr/local/lib/python3.12/dist-packages (from chex>=0.1.87->optax) (0.12.1)
Requirement already satisfied: opt_einsum in /usr/local/lib/python3.12/dist-packages (from jax>=0.5.3->optax) (3.4.0)
Requirement already satisfied: scipy>=1.13 in /usr/local/lib/python3.12/dist-packages (from jax>=0.5.3->optax) (1.16.2)
Requirement already satisfied: markdown-it-py>=2.2.0 in /usr/local/lib/python3.12/dist-packages (from rich->keras) (2.2.0)
Requirement already satisfied: pygments<3.0.0,>=2.13.0 in /usr/local/lib/python3.12/dist-packages (from rich->keras) (2.19.2)
Requirement already satisfied: mdurl~=0.1 in /usr/local/lib/python3.12/dist-packages (from markdown-it-py>=2.2.0->rich->keras) (0.1.2)
os.environ['KERAS_BACKEND'] = 'jax'
Note
Without setting the backend to JAX, the imports below might fail.
If you have problems running the next cell in Jupyter, try
quitting
running
export KERAS_BACKEND="jax"starting Jupyter on the command line from the same terminal.
import keras
from keras import Sequential
from keras.layers import Dense
import optax
23.2. Set Up#
Here we briefly describe the problem and generate synthetic data.
23.2.1. Flow#
We use the routine from Simple Neural Network Regression with Keras and JAX to generate data for one-dimensional nonlinear regression.
Then we will create a dense (i.e., fully connected) neural network with
4 layers, where the input and hidden layers map to
The inputs and outputs are scalar (for one-dimensional nonlinear regression), so the overall mapping is
Here’s a class to store the learning-related constants we’ll use across all implementations.
Our default value of
class Config(NamedTuple):
epochs: int = 4000 # Number of passes through the data set
output_dim: int = 10 # Output dimension of input and hidden layers
learning_rate: float = 0.001 # Learning rate for gradient descent
layer_sizes: tuple = (1, 10, 10, 10, 1) # Sizes of each layer in the network
seed: int = 14 # Random seed for data generation
23.2.2. Data#
Here’s the function to generate the data for our regression analysis.
def generate_data(
key: jax.Array, # JAX random key
data_size: int = 400, # Sample size
x_min: float = 0.0, # Minimum x value
x_max: float = 5.0 # Maximum x value
):
"""
Generate synthetic regression data.
"""
x = jnp.linspace(x_min, x_max, num=data_size)
ϵ = 0.2 * jax.random.normal(key, shape=(data_size,))
y = x**0.5 + jnp.sin(x) + ϵ
# Return observations as column vectors
x = jnp.reshape(x, (data_size, 1))
y = jnp.reshape(y, (data_size, 1))
return x, y
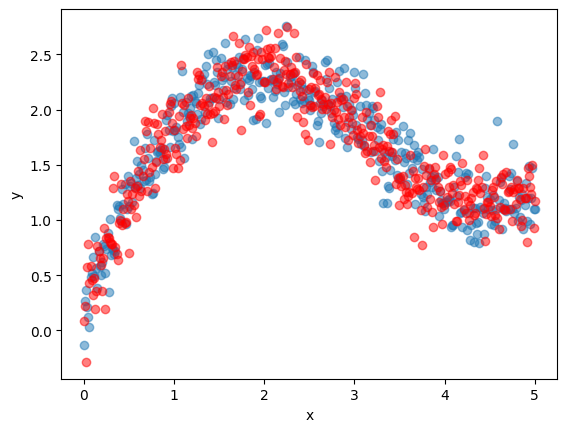
Here’s a plot of the data.
config = Config()
key = jax.random.PRNGKey(config.seed)
key_train, key_validate = jax.random.split(key)
x_train, y_train = generate_data(key_train)
x_validate, y_validate = generate_data(key_validate)
fig, ax = plt.subplots()
ax.scatter(x_train, y_train, alpha=0.5)
ax.scatter(x_validate, y_validate, color='red', alpha=0.5)
ax.set_xlabel('x')
ax.set_ylabel('y')
plt.show()
23.3. Training with Keras#
We build a Keras model that can fit a nonlinear function to the generated data using an ANN.
We will use this fit as a benchmark to test our JAX code.
Since its role is only a benchmark, we refer readers to the previous lecture for details on the Keras interface.
We start with a function to build the model.
def build_keras_model(
config: Config, # contains configuration data
activation_function: str = 'tanh' # activation with default
):
model = Sequential()
# Add layers to the network sequentially, from inputs towards outputs
for i in range(len(config.layer_sizes) - 1):
model.add(
Dense(units=config.output_dim, activation=activation_function)
)
# Add a final layer that maps to a scalar value, for regression.
model.add(Dense(units=1))
# Embed training configurations
model.compile(
optimizer=keras.optimizers.SGD(),
loss='mean_squared_error'
)
return model
Notice that we’ve set the optimizer to use stochastic gradient descent and a mean square loss.
Here is a function to train the model.
def train_keras_model(
model, # Instance of Keras Sequential model
x, # Training data, inputs
y, # Training data, outputs
x_validate, # Validation data, inputs
y_validate, # Validation data, outputs
config: Config # contains configuration data
):
print(f"Training NN using Keras.")
start_time = time()
training_history = model.fit(
x, y,
batch_size=max(x.shape),
verbose=0,
epochs=config.epochs,
validation_data=(x_validate, y_validate)
)
elapsed = time() - start_time
mse = model.evaluate(x_validate, y_validate, verbose=2)
print(f"Trained in {elapsed:.2f} seconds, validation data MSE = {mse}")
return model, training_history, elapsed, mse
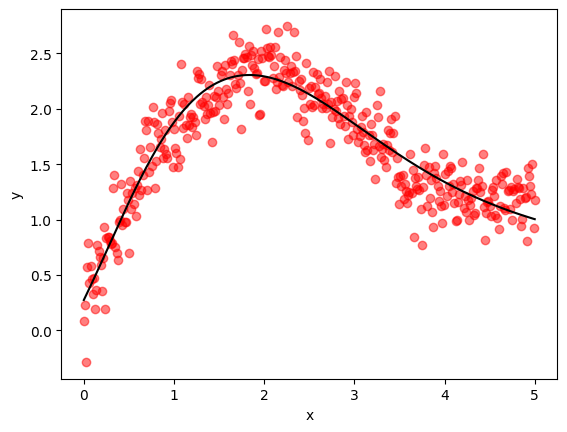
The next function extracts and visualizes a prediction from the trained model.
def plot_keras_output(model, x, y, x_validate, y_validate):
y_predict = model.predict(x_validate, verbose=2)
fig, ax = plt.subplots()
ax.scatter(x_validate, y_validate, color='red', alpha=0.5)
ax.plot(x_validate, y_predict, label="fitted model", color='black')
ax.set_xlabel('x')
ax.set_ylabel('y')
plt.show()
Let’s run the Keras training:
config = Config()
model = build_keras_model(config)
model, training_history, keras_runtime, keras_mse = train_keras_model(
model, x_train, y_train, x_validate, y_validate, config
)
plot_keras_output(model, x_train, y_train, x_validate, y_validate)
The fit is good and we note the relatively low final MSE.
23.4. Training with JAX#
For the JAX implementation, we need to construct the network ourselves, as a map from inputs to outputs.
We’ll use the same network structure we used for the Keras implementation.
23.4.1. Background and set up#
The neural network has the form
Here
Each matrix
The symbol
In fact, when we implement the affine map
the mapping is executed by JAX via the expression
x @ W + b.
Here’s a class to store parameters for one layer of the network.
class LayerParams(NamedTuple):
"""
Stores parameters for one layer of the neural network.
"""
W: jnp.ndarray # weights
b: jnp.ndarray # biases
The following function initializes a single layer of the network using He initialization for weights and ones for biases.
def initialize_layer(in_dim, out_dim, key):
"""
Initialize weights and biases for a single layer of a the network.
Use He initialization for weights and ones for biases.
"""
W = jax.random.normal(key, shape=(in_dim, out_dim)) * jnp.sqrt(2 / in_dim)
b = jnp.ones((1, out_dim))
return LayerParams(W, b)
The next function builds an entire network, as represented by its parameters, by initializing layers and stacking them into a list.
def initialize_network(
key: jax.Array, # JAX random key
config: Config # contains configuration data
):
"""
Build a network by initializing all of the parameters.
A network is a list of LayerParams instances, each
containing a weight-bias pair (W, b).
"""
layer_sizes = config.layer_sizes
params = []
for i in range(len(layer_sizes) - 1):
key, subkey = jax.random.split(key)
layer = initialize_layer(
layer_sizes[i], # in dimension for layer
layer_sizes[i + 1], # out dimension for layer
subkey
)
params.append(layer)
return params
Wait, you say!
Shouldn’t we concatenate the elements of
Actually we don’t need to — we use the JAX PyTree approach discussed below.
23.4.2. Coding the network#
Here’s our implementation of the ANN
def f(
θ: list, # Network parameters (pytree)
x: jnp.ndarray, # Input data (row vector)
σ: callable = jnp.tanh # Activation function
):
"""
Perform a forward pass over the network to evaluate f(θ, x).
"""
*hidden, last = θ
for layer in hidden:
x = σ(x @ layer.W + layer.b)
x = x @ last.W + last.b
return x
The function x and return the predicted vector of outputs y_hat = f(θ, x)
corresponding to each data point.
The loss function is mean squared error, the same as the Keras case.
def loss_fn(
θ: list, # Network parameters (pytree)
x: jnp.ndarray, # Input data
y: jnp.ndarray # Target data
):
return jnp.mean((f(θ, x) - y)**2)
We’ll use its gradient to do stochastic gradient descent.
(Technically, we will be doing gradient descent, rather than stochastic gradient descent, since will not randomize over sample points when we evaluate the gradient.)
loss_gradient = jax.jit(jax.grad(loss_fn))
The gradient of loss_fn is with respect to the first argument θ.
The code above seems kind of magical, since we are differentiating with respect to a parameter “vector” stored as a list of dictionaries containing arrays.
How can we differentiate with respect to such a complex object?
The answer is that the list of dictionaries is treated internally as a pytree.
The JAX function grad understands how to
extract the individual arrays (the “leaves” of the tree),
compute derivatives with respect to each one, and
pack the resulting derivatives into a pytree with the same structure as the parameter vector.
23.4.3. Gradient descent#
Using the above code, we can now write our rule for updating the parameters via gradient descent, which is the algorithm we covered in our lecture on autodiff.
In this case, to keep things as simple as possible, we’ll use a fixed learning rate for every iteration.
def update_parameters(
θ: list, # Current parameters (pytree)
x: jnp.ndarray, # Input data
y: jnp.ndarray, # Target data
config: Config # contains configuration data
):
"""
Update the parameter pytree using gradient descent.
"""
λ = config.learning_rate
# Specify the update rule
def gradient_descent_step(p, g):
"""
A rule for updating parameter vector p given gradient vector g.
It will be applied to each leaf of the pytree of parameters.
"""
return p - λ * g
gradient = loss_gradient(θ, x, y)
# Use tree.map to apply the update rule to the parameter vectors
θ_new = jax.tree.map(gradient_descent_step, θ, gradient)
return θ_new
Here jax.tree.map understands θ and gradient as pytrees of the
same structure and executes p - λ * g on the corresponding leaves of the pair
of trees.
Each weight matrix and bias vector is updated by gradient descent, exactly as required.
Here’s code that puts this all together.
@partial(jax.jit, static_argnames=['config'])
def train_jax_model(
θ: list, # Initial parameters (pytree)
x: jnp.ndarray, # Training input data
y: jnp.ndarray, # Training target data
config: Config # contains configuration data
):
"""
Train model using gradient descent.
"""
def update(_, θ):
θ_new = update_parameters(θ, x, y, config)
return θ_new
θ_final = jax.lax.fori_loop(0, config.epochs, update, θ)
return θ_final
23.4.4. Execution#
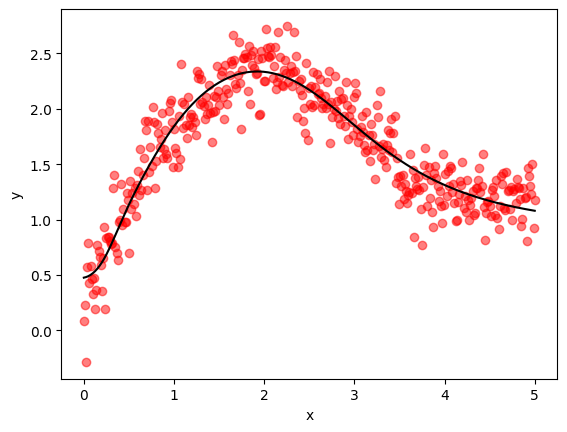
Let’s run our code and see how it goes.
We’ll reuse the data we generated earlier.
# Reset parameter vector
config = Config()
param_key = jax.random.PRNGKey(1234)
θ = initialize_network(param_key, config)
# Warmup run to trigger JIT compilation
train_jax_model(θ, x_train, y_train, config)
# Reset and time the actual run
θ = initialize_network(param_key, config)
start_time = time()
θ = train_jax_model(θ, x_train, y_train, config)
θ[0].W.block_until_ready() # Ensure computation completes
jax_runtime = time() - start_time
jax_mse = loss_fn(θ, x_validate, y_validate)
jax_train_mse = loss_fn(θ, x_train, y_train)
print(f"Trained model with JAX in {jax_runtime:.2f} seconds.")
print(f"Final MSE on validation data = {jax_mse:.6f}")
Trained model with JAX in 0.27 seconds.
Final MSE on validation data = 0.042119
Despite the simplicity of our implementation, we actually perform slightly better than Keras.
Here’s a visualization of the quality of our fit.
23.5. JAX plus Optax#
Our hand-coded optimization routine above was quite effective, but in practice we might wish to use an optimization library written for JAX.
One such library is Optax.
23.5.1. Optax with SGD#
Here’s a training routine using Optax’s stochastic gradient descent solver.
@partial(jax.jit, static_argnames=['config'])
def train_jax_optax(
θ: list, # Initial parameters (pytree)
x: jnp.ndarray, # Training input data
y: jnp.ndarray, # Training target data
config: Config # contains configuration data
):
" Train model using Optax SGD optimizer. "
epochs = config.epochs
learning_rate = config.learning_rate
solver = optax.sgd(learning_rate)
opt_state = solver.init(θ)
def update(_, loop_state):
θ, opt_state = loop_state
grad = loss_gradient(θ, x, y)
updates, new_opt_state = solver.update(grad, opt_state, θ)
θ_new = optax.apply_updates(θ, updates)
new_loop_state = θ_new, new_opt_state
return new_loop_state
initial_loop_state = θ, opt_state
final_loop_state = jax.lax.fori_loop(0, epochs, update, initial_loop_state)
θ_final, _ = final_loop_state
return θ_final
Let’s try running it.
# Reset parameter vector
θ = initialize_network(param_key, config)
# Warmup run to trigger JIT compilation
train_jax_optax(θ, x_train, y_train, config)
# Reset and time the actual run
θ = initialize_network(param_key, config)
start_time = time()
θ = train_jax_optax(θ, x_train, y_train, config)
θ[0].W.block_until_ready() # Ensure computation completes
optax_sgd_runtime = time() - start_time
optax_sgd_mse = loss_fn(θ, x_validate, y_validate)
optax_sgd_train_mse = loss_fn(θ, x_train, y_train)
print(f"Trained model with JAX and Optax SGD in {optax_sgd_runtime:.2f} seconds.")
print(f"Final MSE on validation data = {optax_sgd_mse:.6f}")
Trained model with JAX and Optax SGD in 0.27 seconds.
Final MSE on validation data = 0.042119
23.5.2. Optax with ADAM#
We can also consider using a slightly more sophisticated gradient-based method, such as ADAM.
You will notice that the syntax for using this alternative optimizer is very similar.
@partial(jax.jit, static_argnames=['config'])
def train_jax_optax_adam(
θ: list, # Initial parameters (pytree)
x: jnp.ndarray, # Training input data
y: jnp.ndarray, # Training target data
config: Config # contains configuration data
):
" Train model using Optax ADAM optimizer. "
epochs = config.epochs
learning_rate = config.learning_rate
solver = optax.adam(learning_rate)
opt_state = solver.init(θ)
def update(_, loop_state):
θ, opt_state = loop_state
grad = loss_gradient(θ, x, y)
updates, new_opt_state = solver.update(grad, opt_state, θ)
θ_new = optax.apply_updates(θ, updates)
return (θ_new, new_opt_state)
initial_loop_state = θ, opt_state
θ_final, _ = jax.lax.fori_loop(0, epochs, update, initial_loop_state)
return θ_final
# Reset parameter vector
θ = initialize_network(param_key, config)
# Warmup run to trigger JIT compilation
train_jax_optax_adam(θ, x_train, y_train, config)
# Reset and time the actual run
θ = initialize_network(param_key, config)
start_time = time()
θ = train_jax_optax_adam(θ, x_train, y_train, config)
θ[0].W.block_until_ready() # Ensure computation completes
optax_adam_runtime = time() - start_time
optax_adam_mse = loss_fn(θ, x_validate, y_validate)
optax_adam_train_mse = loss_fn(θ, x_train, y_train)
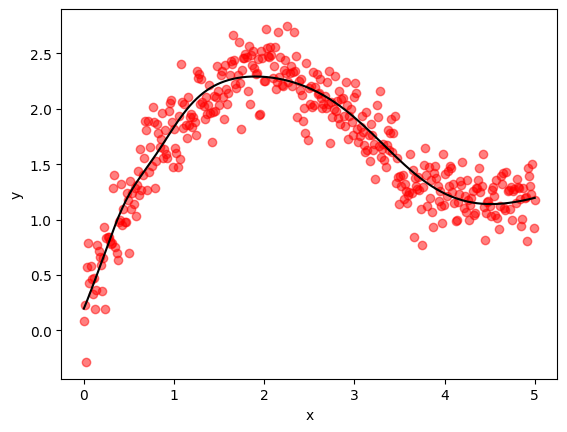
print(f"Trained model with JAX and Optax ADAM in {optax_adam_runtime:.2f} seconds.")
print(f"Final MSE on validation data = {optax_adam_mse:.6f}")
Trained model with JAX and Optax ADAM in 0.28 seconds.
Final MSE on validation data = 0.040869
Here’s a visualization of the result.
23.6. Summary#
Here we compare the performance of the four different training approaches we explored in this lecture.
Show code cell source
import pandas as pd
# Compute training MSEs for each method
# Need to retrieve the trained models and compute training MSE
# For Keras, we already have the model
keras_train_mse = model.evaluate(x_train, y_train, verbose=0)
# For JAX methods, we need to compute using loss_fn with the final θ from each method
# We need to re-train or save the θ from each method
# For now, let's add these calculations after each training section
# Create summary table
results = {
'Method': [
'Keras',
'Pure JAX (hand-coded GD)',
'JAX + Optax SGD',
'JAX + Optax ADAM'
],
'Runtime (s)': [
keras_runtime,
jax_runtime,
optax_sgd_runtime,
optax_adam_runtime
],
'Training MSE': [
keras_train_mse,
jax_train_mse,
optax_sgd_train_mse,
optax_adam_train_mse
],
'Validation MSE': [
keras_mse,
jax_mse,
optax_sgd_mse,
optax_adam_mse
]
}
df = pd.DataFrame(results)
# Format MSE columns to 6 decimal places
df['Training MSE'] = df['Training MSE'].apply(lambda x: f"{x:.6f}")
df['Validation MSE'] = df['Validation MSE'].apply(lambda x: f"{x:.6f}")
print("\nSummary of Training Methods:")
print(df.to_string(index=False))
Summary of Training Methods:
Method Runtime (s) Training MSE Validation MSE
Keras 18.769433 0.040176 0.044817
Pure JAX (hand-coded GD) 0.273956 0.039963 0.042119
JAX + Optax SGD 0.273528 0.039963 0.042119
JAX + Optax ADAM 0.277265 0.035879 0.040869
All methods achieve similar validation MSE values (around 0.043-0.045).
At the time of writing, the MSEs from plain vanilla Optax and our own hand-coded SGD routine are identical.
The ADAM optimizer achieves slightly better MSE by using adaptive learning rates.
Still, our hand-coded algorithm does very well compared to this high-quality optimizer.
Note also that the pure JAX implementations are significantly faster than Keras.
This is because JAX can JIT-compile the entire training loop.
Not surprisingly, Keras has more overhead from its abstraction layers.
23.7. Exercises#
Exercise 23.1
Try to reduce the MSE on the validation data without significantly increasing the computational load.
You should hold constant both the number of epochs and the total number of parameters in the network.
Currently, the network has 4 layers with output dimension
Layer 0:
Layer 1:
Layer 2:
Layer 3:
Total:
You can experiment with:
Changing the network architecture
Trying different activation functions (e.g.,
jax.nn.relu,jax.nn.gelu,jax.nn.sigmoid,jax.nn.elu)Modifying the optimizer (e.g., different learning rates, learning rate schedules, momentum, other Optax optimizers)
Experimenting with different weight initialization strategies
Which combination gives you the lowest validation MSE?
Solution to Exercise 23.1
Let’s implement and test several strategies.
Strategy 1: Deeper Network Architecture
Let’s try a deeper network with 6 layers instead of 4, keeping total parameters ≤ 251:
# Strategy 1: Deeper network (6 layers with k=6)
# Layer sizes: 1→6→6→6→6→6→1
# Parameters: (1×6+6) + 4×(6×6+6) + (6×1+1) = 12 + 4×42 + 7 = 187 < 251
θ = initialize_network(param_key, config)
def initialize_deep_params(
key: jax.Array, # JAX random key
k: int = 6, # Layer width
num_hidden: int = 5 # Number of hidden layers
):
" Initialize parameters for deeper network with k=6. "
layer_sizes = tuple([1] + [k] * num_hidden + [1])
config_deep = Config(layer_sizes=layer_sizes)
return initialize_network(key, config_deep)
θ_deep = initialize_deep_params(param_key)
config_deep = Config(layer_sizes=(1, 6, 6, 6, 6, 6, 1))
# Warmup
train_jax_optax_adam(θ_deep, x_train, y_train, config_deep)
# Actual run
θ_deep = initialize_deep_params(param_key)
start_time = time()
θ_deep = train_jax_optax_adam(θ_deep, x_train, y_train, config_deep)
θ_deep[0].W.block_until_ready()
deep_runtime = time() - start_time
deep_mse = loss_fn(θ_deep, x_validate, y_validate)
print(f"Strategy 1 - Deeper network (6 layers, k=6)")
print(f" Total parameters: 187")
print(f" Runtime: {deep_runtime:.2f}s")
print(f" Validation MSE: {deep_mse:.6f}")
print(f" Improvement over ADAM: {optax_adam_mse - deep_mse:.6f}")
Strategy 1 - Deeper network (6 layers, k=6)
Total parameters: 187
Runtime: 0.46s
Validation MSE: 0.041686
Improvement over ADAM: -0.000817
Strategy 2: Deeper Network + Learning Rate Schedule
Since the deeper network performed best, let’s combine it with the learning rate schedule:
# Strategy 2: Deeper network + LR schedule
θ_deep = initialize_deep_params(param_key)
def train_deep_with_schedule(
θ: list,
x: jnp.ndarray,
y: jnp.ndarray,
config: Config
):
" Train deeper network with learning rate schedule. "
epochs = config.epochs
schedule = optax.exponential_decay(
init_value=0.003,
transition_steps=1000,
decay_rate=0.5
)
solver = optax.adam(schedule)
opt_state = solver.init(θ)
def update(_, loop_state):
θ, opt_state = loop_state
grad = loss_gradient(θ, x, y)
updates, new_opt_state = solver.update(grad, opt_state, θ)
θ_new = optax.apply_updates(θ, updates)
return (θ_new, new_opt_state)
initial_loop_state = θ, opt_state
θ_final, _ = jax.lax.fori_loop(0, epochs, update, initial_loop_state)
return θ_final
# Warmup
train_deep_with_schedule(θ_deep, x_train, y_train, config_deep)
# Actual run
θ_deep = initialize_deep_params(param_key)
start_time = time()
θ_deep_schedule = train_deep_with_schedule(θ_deep, x_train, y_train, config_deep)
θ_deep_schedule[0].W.block_until_ready()
deep_schedule_runtime = time() - start_time
deep_schedule_mse = loss_fn(θ_deep_schedule, x_validate, y_validate)
print(f"Strategy 2 - Deeper network + LR schedule")
print(f" Runtime: {deep_schedule_runtime:.2f}s")
print(f" Validation MSE: {deep_schedule_mse:.6f}")
print(f" Improvement over ADAM: {optax_adam_mse - deep_schedule_mse:.6f}")
Strategy 2 - Deeper network + LR schedule
Runtime: 1.02s
Validation MSE: 0.041633
Improvement over ADAM: -0.000764
Strategy 3: Deeper Network + LR Schedule + L2 Regularization
Let’s add L2 regularization (similar to ridge regression) to penalize complexity:
# Strategy 3: Deeper network + LR schedule + L2 regularization
θ_deep = initialize_deep_params(param_key)
def train_deep_with_schedule_and_l2(
θ: list,
x: jnp.ndarray,
y: jnp.ndarray,
config: Config,
lambda_l2: float = 0.001
):
" Train deeper network with learning rate schedule and L2 regularization. "
epochs = config.epochs
schedule = optax.exponential_decay(
init_value=0.003,
transition_steps=1000,
decay_rate=0.5
)
# Define regularized loss function
@jax.jit
def loss_fn_l2(θ, x, y):
# Standard MSE loss
mse = jnp.mean((f(θ, x) - y)**2)
# L2 penalty on weights (not biases)
l2_penalty = 0.0
for W, b in θ:
l2_penalty += jnp.sum(W**2)
return mse + lambda_l2 * l2_penalty
loss_gradient_l2 = jax.jit(jax.grad(loss_fn_l2))
solver = optax.adam(schedule)
opt_state = solver.init(θ)
def update(_, loop_state):
θ, opt_state = loop_state
grad = loss_gradient_l2(θ, x, y)
updates, new_opt_state = solver.update(grad, opt_state, θ)
θ_new = optax.apply_updates(θ, updates)
return (θ_new, new_opt_state)
initial_loop_state = θ, opt_state
θ_final, _ = jax.lax.fori_loop(0, epochs, update, initial_loop_state)
return θ_final
# Warmup
train_deep_with_schedule_and_l2(θ_deep, x_train, y_train, config_deep)
# Actual run
θ_deep = initialize_deep_params(param_key)
start_time = time()
θ_deep_l2 = train_deep_with_schedule_and_l2(θ_deep, x_train, y_train, config_deep)
θ_deep_l2[0].W.block_until_ready()
deep_l2_runtime = time() - start_time
deep_l2_mse = loss_fn(θ_deep_l2, x_validate, y_validate)
print(f"Strategy 3 - Deeper network + LR schedule + L2 regularization")
print(f" Runtime: {deep_l2_runtime:.2f}s")
print(f" Validation MSE: {deep_l2_mse:.6f}")
print(f" Improvement over ADAM: {optax_adam_mse - deep_l2_mse:.6f}")
Strategy 3 - Deeper network + LR schedule + L2 regularization
Runtime: 1.15s
Validation MSE: 0.040845
Improvement over ADAM: 0.000024
Strategy 4: Baseline + L2 Regularization
Let’s see if L2 regularization helps the baseline architecture:
# Strategy 4: Baseline architecture + L2 regularization
θ = initialize_network(param_key, config)
def train_baseline_with_l2(
θ: list,
x: jnp.ndarray,
y: jnp.ndarray,
config: Config,
lambda_l2: float = 0.001
):
" Train baseline model with L2 regularization. "
epochs = config.epochs
learning_rate = config.learning_rate
# Define regularized loss function
@jax.jit
def loss_fn_l2(θ, x, y):
# Standard MSE loss
mse = jnp.mean((f(θ, x) - y)**2)
# L2 penalty on weights (not biases)
l2_penalty = 0.0
for W, b in θ:
l2_penalty += jnp.sum(W**2)
return mse + lambda_l2 * l2_penalty
loss_gradient_l2 = jax.jit(jax.grad(loss_fn_l2))
solver = optax.adam(learning_rate)
opt_state = solver.init(θ)
def update(_, loop_state):
θ, opt_state = loop_state
grad = loss_gradient_l2(θ, x, y)
updates, new_opt_state = solver.update(grad, opt_state, θ)
θ_new = optax.apply_updates(θ, updates)
return (θ_new, new_opt_state)
initial_loop_state = θ, opt_state
θ_final, _ = jax.lax.fori_loop(0, epochs, update, initial_loop_state)
return θ_final
# Warmup
train_baseline_with_l2(θ, x_train, y_train, config)
# Actual run
θ = initialize_network(param_key, config)
start_time = time()
θ_baseline_l2 = train_baseline_with_l2(θ, x_train, y_train, config)
θ_baseline_l2[0].W.block_until_ready()
baseline_l2_runtime = time() - start_time
baseline_l2_mse = loss_fn(θ_baseline_l2, x_validate, y_validate)
print(f"Strategy 4 - Baseline + L2 regularization")
print(f" Runtime: {baseline_l2_runtime:.2f}s")
print(f" Validation MSE: {baseline_l2_mse:.6f}")
print(f" Improvement over ADAM: {optax_adam_mse - baseline_l2_mse:.6f}")
Strategy 4 - Baseline + L2 regularization
Runtime: 0.84s
Validation MSE: 0.040959
Improvement over ADAM: -0.000090
Results Summary
Let’s compare all strategies:
Show code cell source
# Summary of all strategies
strategies_results = {
'Strategy': [
'Baseline (ADAM + tanh)',
'1. Deeper network (6 layers)',
'2. Deeper network + LR schedule',
'3. Strategy 2 + L2 regularization',
'4. Baseline + L2 regularization'
],
'Runtime (s)': [
optax_adam_runtime,
deep_runtime,
deep_schedule_runtime,
deep_l2_runtime,
baseline_l2_runtime
],
'Validation MSE': [
optax_adam_mse,
deep_mse,
deep_schedule_mse,
deep_l2_mse,
baseline_l2_mse
],
'Improvement': [
0.0,
float(optax_adam_mse - deep_mse),
float(optax_adam_mse - deep_schedule_mse),
float(optax_adam_mse - deep_l2_mse),
float(optax_adam_mse - baseline_l2_mse)
]
}
df_strategies = pd.DataFrame(strategies_results)
print("\nSummary of Exercise Strategies:")
print(df_strategies.to_string(index=False))
Summary of Exercise Strategies:
Strategy Runtime (s) Validation MSE Improvement
Baseline (ADAM + tanh) 0.277265 0.0408687 0.000000
1. Deeper network (6 layers) 0.459583 0.041686147 -0.000817
2. Deeper network + LR schedule 1.016304 0.041632924 -0.000764
3. Strategy 2 + L2 regularization 1.147835 0.040844545 0.000024
4. Baseline + L2 regularization 0.835761 0.04095904 -0.000090
The experimental results reveal several lessons:
Architecture matters: A deeper, narrower network outperformed the baseline network, despite using fewer parameters (187 vs 251).
Combining strategies: Combining the deeper architecture with a learning rate schedule showed that synergistic improvements are possible.
Regularization helps: Adding L2 regularization (ridge penalty) can improve performance by penalizing model complexity and reducing overfitting.
Regularization vs architecture: Comparing strategies 3 and 4 shows whether regularization is more effective with deeper architectures or simpler ones.